1. Site-specific protein labeling and cell imaging.
Virtually all the biological processes inside the living cell are dependent on proteins and their interaction networks. Protein labeling plays a key role for understanding the biological function of a protein because it enables us to track the production, distribution, translocation and degradation of a specific protein and follow its interaction with other proteins inside a living cell. We recently developed an efficient protein labeling method based on the protein posttranslational modification reaction catalyzed by phosphopantetheinyl transferases. This protein labeling method has the combined advantage of: (1) small size of the peptide tags for the construction of protein fusions with the minimum disturbance to the target protein structure and biological function; (2) one step protein labeling for the direct conjugation of small molecule probes of diverse structures to the tagged target protein; (3) high efficiency and specificity of the labeling reaction that is compatible for living cell imaging. We are now using this method to label cell membrane proteins for the elucidation of their biological functions. We are also trying to adapt this method for the labeling of intracellular proteins.
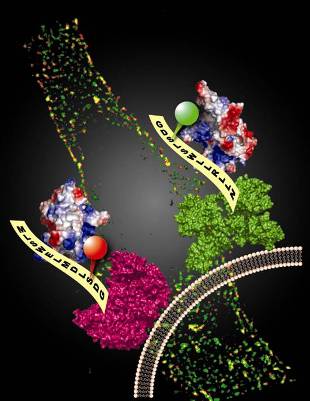
Orthogonal protein labeling by phosphopantetheinyl transferases.
2. Enzyme evolution for the combinatorial biosynthesis of natural products.
Natural products possess enormous structural and chemical diversities and have been the best source of bioactive compounds that supplies the need of the drug discovery pipeline. However natural product based drug discovery is hampered by the dilemma that though the structural complexity of the natural product molecules gives rise to compounds with diverse biological activities as promising drug leads, it also presents daunting challenges for modifying the natural product molecules by chemical synthesis for the optimization of their medicinal activity. We are in the process to evolve the substrate specificity of the enzymatic modules in natural product biosynthetic enzymes for the incorporation of structurally diversified building blocks into their final structure. In this way libraries of “unnatural” natural products can be generated biosynthetically, bypassing the hurdle in the total synthesis of the natural product analogs.
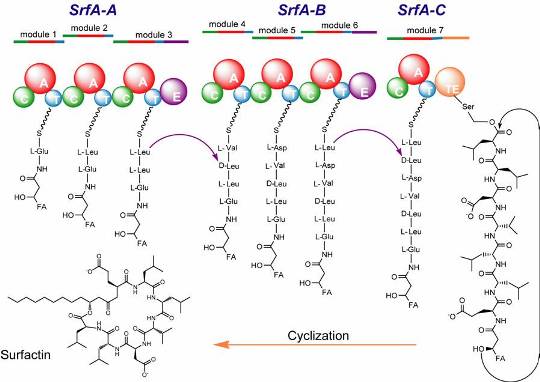
A natural product biosynthetic assembly line for protein engineering and enzyme evolution.
3. Profiling posttranslational modification enzymes mediating signal transduction.
Communication and coordination of various cellular events from cell division to apoptosis depend on the transduction of dynamic cellular signals encoded by one universal chemical language – protein posttranslational modification (PTM). For example, PTMs such as phosphorylation, glycosylation, methylation, acetylation and lipidation all play central roles mediating signal transduction either by modulating protein – protein interactions or by affecting the subcellular localization of the modified proteins. Two major challenges for mapping the signal transduction networks are: (1) how to identify from the whole proteome the downstream targets of a PTM enzyme such as a kinase or a glycosyltransferase and (2) how to identify from the whole proteome the upstream PTM enzymes that are responsible for a specific modification such as phosphorylation or glycosylation on a signaling molecule. We are thus interested in developing high throughput profiling systems that would allow us to profile the PTM enzymes in the human genome and address those challenges.
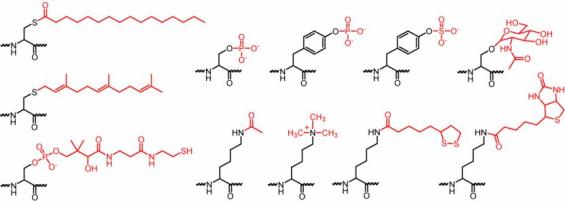
Various protein posttranslational modifications.